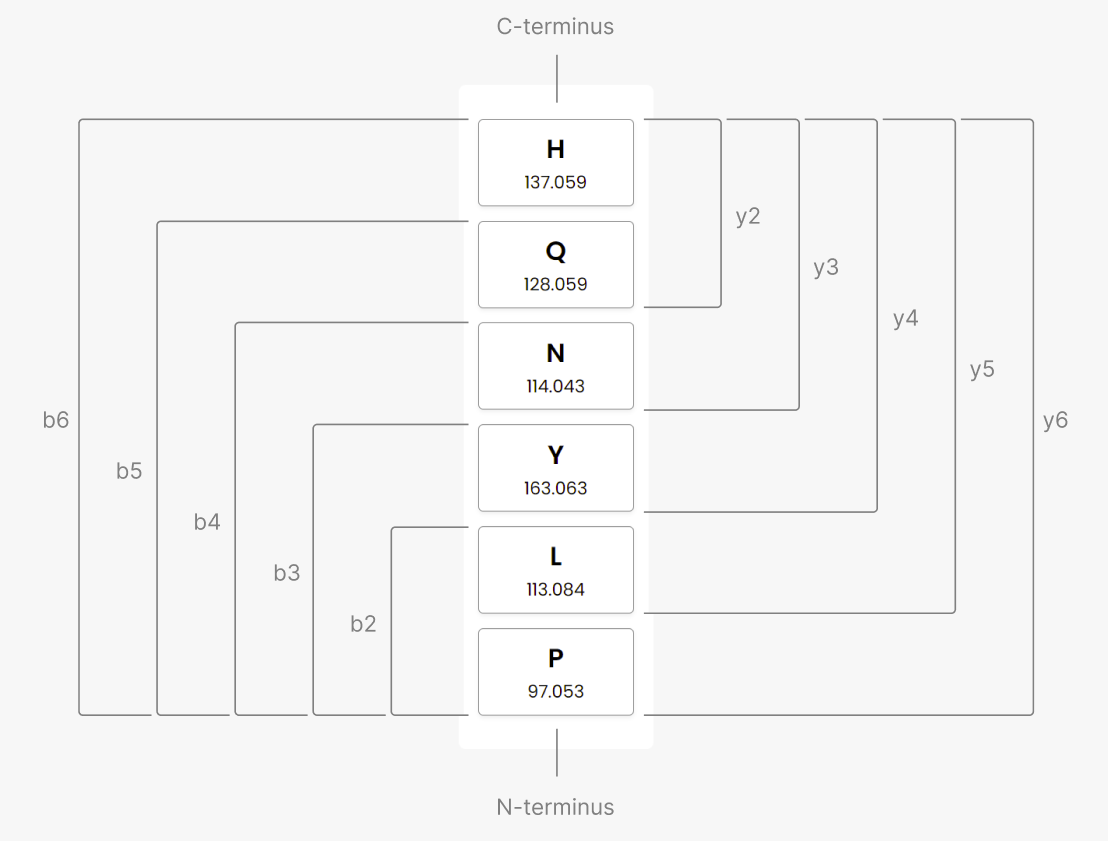
Peptide Sequencing Simulation
This is a teaching tool demonstrating the relationship between the order of amino acids in a peptide and the peptide’s mass spectrometry data. The list on the left represents a sequence of amino acids forming a peptide, and the graphs on the right represent simulated mass spectrometry data from that peptide. Rearranging the peptide changes the simulated masses shown in the "Current Masses" graph. Based on the masses shown in the "Goal Masses" graph, reorder the "Current" amino acids to match the peptide.
- Drag and drop an amino acid cell to rearrange the peptide.
- Hover over the bars to reveal the combined masses.
- Click a bar to highlight the fragment it corresponds to.